NOTOS measurements are usually done on 13 and/or 5 mm diameter pellets (XAS) and capillaries (XRD/XAS). Please calculate the linear absorption coefficient μ before preparing your sample. In the case of XAS measurement, we recommend not exceeding 1.5 cm-1 total absorption to avoid distortion and dumping of the EXAFS oscillations. For XRD calculate the relative absorption (μr) at the wavelength (energy) at which patterns will be collected. Note, to minimize absorption correction μr should be <1 (no correction needed) and never higher than 4.5. μr must take into account the packing density (packing fraction, generally 40-60%).
The following web pages and program provide tools to calculate μ:
- XAFSmass — XAFSmassQt 1.5.0 documentation
- 11-BM Resources (X-ray Absorption) (anl.gov)
- NIST X-Ray Form Factor, Atten. Scatt. Tables Form Page
Quartz and borosilicate capillaries of diameter 0.5, 0.7, 1.0, and 1.5 mm are commonly used at the beamline. Please consider also the wall absorption when preparing your samples:
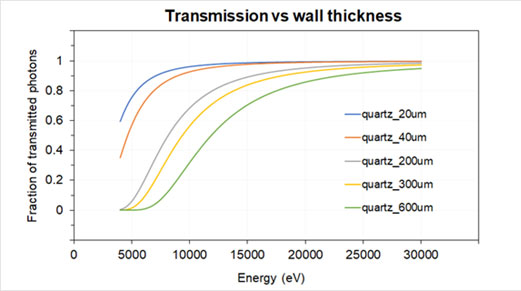
As a general policy, users need to bring and use their own capillaries to run the experiment. Capillaries can be ordered from several companies like Hilgenberg, WJM-Glas Müller or CTS Capillary Tube Supplies.
The same policy applies for the use of coin cells in electrochemistry experiments. To order your coin cells, please contact Michael Knapp and Almuth Pfleging.
In case of necessity, NOTOS counts on a minimum storage of capillaries and coin cells. In the case of use during your experiment, you need to return number of pieces used in the shortest time.
Please contact the local contact and try to coordinate with her/him to avoid any change of setup and/or configuration outside the standard working hours.
Data treatment on the beamline can be performed using standard programs like:
- Athena (https://bruceravel.github.io/demeter/)
- GSAS (https://www.aps.anl.gov/Science/Scientific-Software/GSASII)
while a fast data reduction and visualization can be done using the Fast data Analyser (FDA) a Python utility written by C. Marini, oriented to the analysis of X-ray absorption spectra and XRD patterns. A paper is in preparation. The latest version of the code (0.9.3) can be downloaded at this link: https://git.cells.es/cmarini/fda_free/-/blob/master/fda_free.093.7z . Please read the file "README.TXT" to install and run the program.